Chorus2
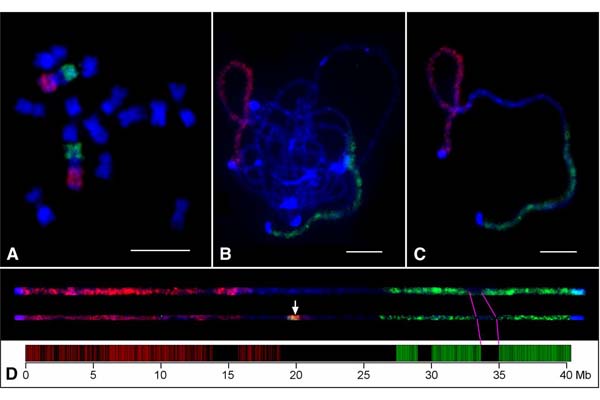
Design of genome-scale oligonucleotide-based probes for fluorescence in situ hybridization (FISH)
Introduction
Chorus2, a software that is developed specifically for oligo selection in plant species. It is highly effective to remove all repetitive elements in selection of single copy oligos, which is critical for the development of successful FISH probes. Chorus2 is more effective than Chorus and OligoMiner for repeat removal. Chorus2 allows to select oligos that are conserved among related species, which extends the usage of oligo-FISH probes among phylogenetically related plant species. Chorus2 allows development of FISH probes from plant species without an assembled genome.
Video tutorials
Oligo design with Chorus/Chorus2
- Wheat: Li GR†, Zhang T†, Yu ZH, Wang HJ, Yang EN, Yang ZJ*. An efficient Oligo‐FISH painting system for revealing chromosome rearrangements and polyploidization in Triticeae. The Plant Journal 2021, 105(4):978-993
- Rice: Liu XY†, Sun S†, Wu Y, Zhou Y, Gu S.W, Yu H.X, Yi CD, Gu MH, Jiang JM, Liu B, Zhang T*, Gong ZY*. Dual‐color oligo‐FISH can reveal chromosomal variations and evolution in Oryza species. The Plant Journal 2020, 101:112-121
- Maize: Albert P.S†, Zhang T†, Semrau K, Rouillard JM, Kao YH, Wang CJ, Danilova T.V, Jiang JM, and Birchler J*. Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships Proceedings of the National Academy of Sciences of the United States of America 2019, 16(5):1679-1685
- Populus: Xin HY†, Zhang T†, Wu YF, Zhang WL, Zhang PD, Xi ML*, Jiang JM. An extraordinarily stable karyotype of the woody Populus species revealed by chromosome painting. The Plant Journal 2020, 101:253-264
Xin H†, Zhang T†, Han Y, Wu YF, Shi JS, Xi ML*, and Jiang JM. Chromosome painting and comparative physical mapping of the sex chromosomes in Populus tomentosa and Populus deltoides. Chromosoma 2018, 127(3):313-321 - Cotton: Liu YL, Wang XY, Wei YY, Liu Z, Lu QW, Liu F, Zhang T*, Peng RH*. Chromosome Painting Based on Bulked Oligonucleotides in Cotton Frontier in Plant Science 2020, 11:802
- Sugarcane: Zhang Q†, Qi YY†, Pan HR†, Tang HB, Wang G, Hua XT, Wang YJ, Lin LY, Li Z, Li YH, Yu F, Yu ZH, Huang YJ, Wang TY, Ma PP, Dou MJ, Sun ZY, Wang YB, Wang HB, Zhang XT, Yao W, Wang YT, Liu XL, Wang MJ, Wang JP, Deng ZH, Xu JS, Yang QH, Liu ZJ, Chen BS, Zhang MQ, Ming R, Zhang JS*. Genomic insights into the recent chromosome reduction of autopolyploid sugarcane Saccharum spontaneum Nature Genetics 2022, 54:885-896
- Potato, Tomato, Eggplant, Tzimbalo and Pepper: Braz GT†, He L†, Zhao HN†, Zhang T†, Semrau K, Rouillard JM, Torres GA, Jiang JM*. Comparative oligo-FISH mapping: an efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics 2018, 208(2):513-523
- Cucumber, Muskmelon and other Cucumis: Han YH†, Zhang T†, Thammapichai P, Weng YQ, and Jiang JM*. Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 2015, 200(3):771-779
- Chicken: Huang Z†, Xu ZX†, Bai H†, Huang YJ, Kang N, Ding XT, Liu J, Luo HR, Yang CT, Chen WJ, Guo QX, XUE LZ, ZhangXP, Xu L, Chen ML, Fu HG, Chen YL, Yue ZC, Fukagawa T, Liu SL, Chang GB*, Xu LH*. Evolutionary analysis of a complete chicken genome Proceedings of the National Academy of Sciences of the United States of America 2023, 120(8)e2216641120
- Switchgrass, Banana and etc …
Oligo-FISH performed with Chorus/Chorus2 designed probes
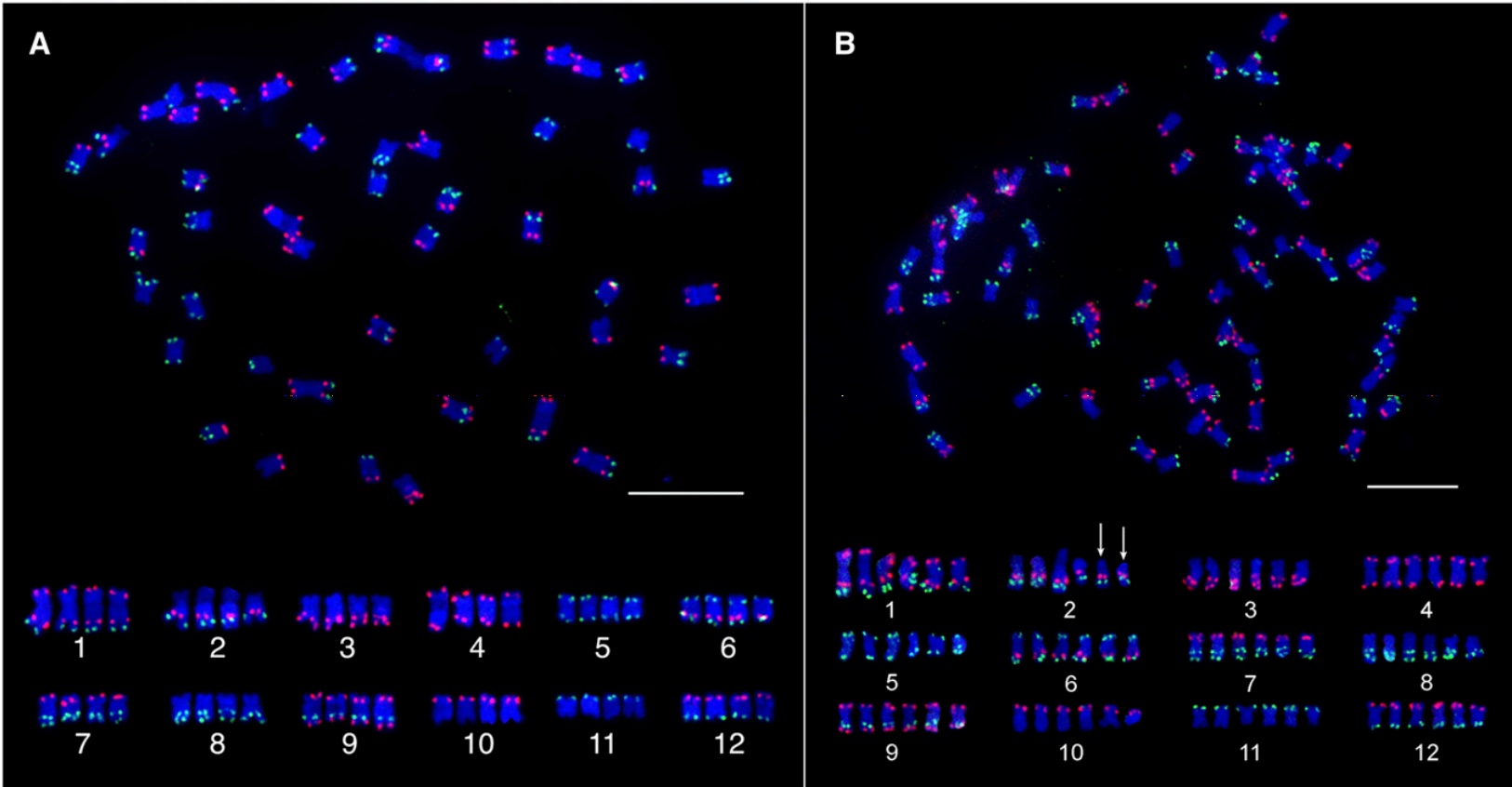
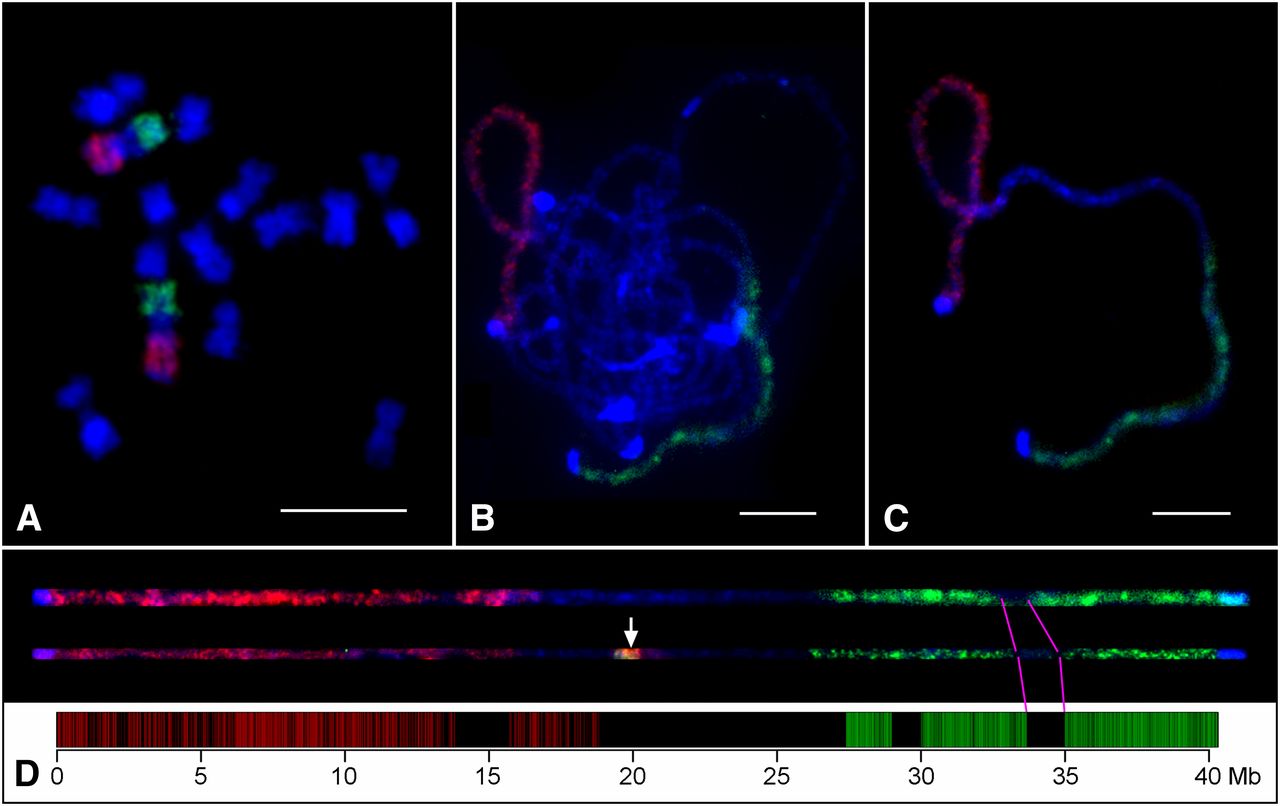
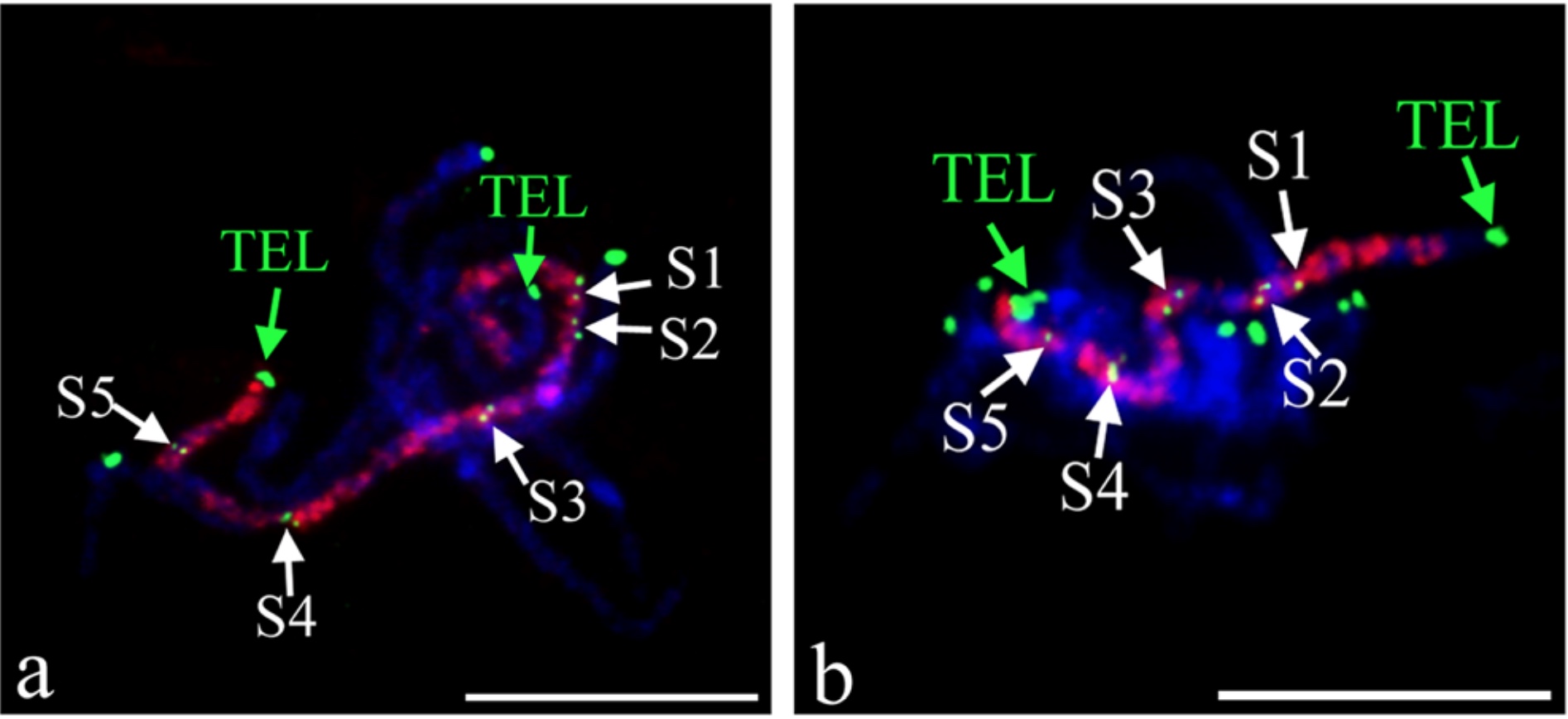
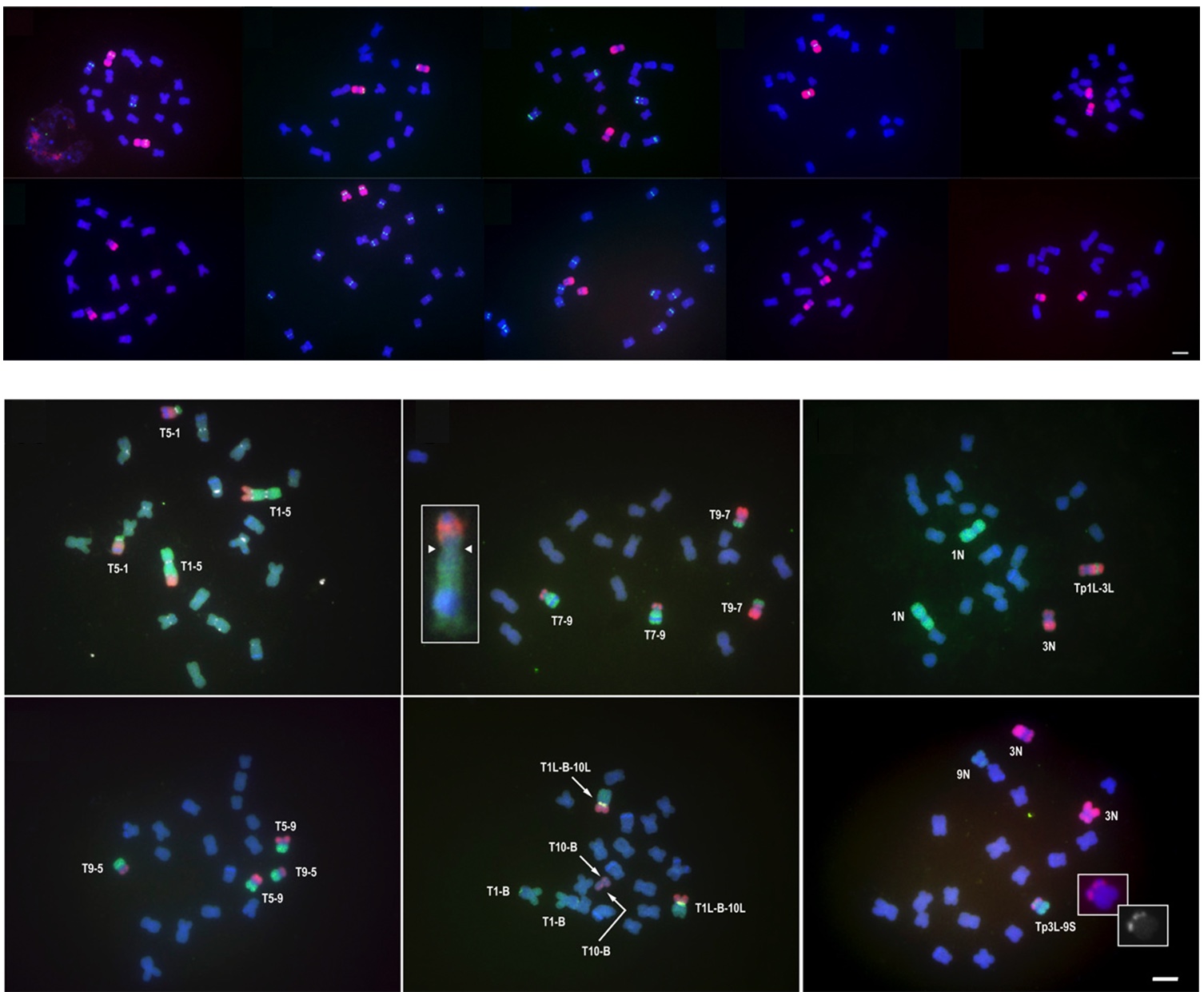
Citation
Zhang T†,*, Liu GQ†, Zhao HN, Braz G.T, Jiang JM*. Chorus2: design of genome-scale oligonucleotide-based probes for fluorescence in situ hybridization Plant Biotechnology Journal (2021) 2021, 19(10):1967-1978
Oligo datasets
We have developed oligo datasets for several plant species. The datasets currently cover six popular plant species.
The oligo datasets is available here.